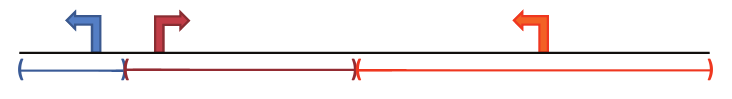
Association Rules
GREAT calculates statistics by associating genomic regions with nearby genes and applying the gene annotations to the regions. Association is a two-step process. First, each gene is assigned a regulatory domain (i.e. a genomic range wherein regulatory elements are likely to target the gene). Then, a genomic region is associated with all genes whose regulatory region it overlaps. Please also see Genes page for the list of genes considered in GREAT.
How does GREAT define gene regulatory domains?
As of yet, there are no clear methods for identifying the regulatory domain of a gene. It is known that regulatory elements can be located over one million bases away from their target genes and even jump over intervening genes. GREAT allows you to choose from among three approaches to defining gene regulatory domains.
Note that GREAT measures all distances from the transcription start site of a gene's canonical isoform.
Approach 1 - Basal plus extension
Each gene is assigned a basal regulatory domain of a minimum distance upstream and downstream of the TSS (regardless of other nearby genes). The gene regulatory domain is extended in both directions to the nearest gene's basal domain but no more than a maximum extension in one direction. When extending the regulatory domain of gene G beyond its basal domain, the extension to the "left" extends until it reaches the first basal domain of any gene whose transcription start site is "left" of G's transcription start site (and analogously for extending "right").
Note: The methodology of extending beyond basal domains was slightly different in the previous GREAT v1.2 release.
Approach 2 - Two nearest genes
Each gene is assigned a regulatory domain that extends in both directions to the nearest gene's TSS but no more than a maximum extension in one direction.
Approach 3 - Single nearest gene
Each gene is assigned a regulatory domain that extends in both directions to the midpoint between the gene's TSS and the nearest gene's TSS but no more than a maximum extension in one direction. This has the effect of assigning a region to the single nearest gene.
All three of the regulatory domain assignment rules are based on a unique ordering of the loci within the genome. However, there are a few distinct loci that possess identical canonical TSS positions. GREAT breaks ties in TSS positions using the arbitrary comparison of strand (+ first) and then the UCSC ID of the gene.
Curated Regulatory Domains
In addition to the above association rules, GREAT utilizes a set of literature curated regulatory domains. Where experimental evidence demonstrates that a gene is directly regulated by an element that falls outside of its putative regulatory domain (as defined by the default Basal plus extension rule), GREAT includes a curated regulatory domain that extends the regulatory domain for the gene to include its known regulatory element. The curated regulatory domains override the automatic rules. To ignore the curated regulatory domains, uncheck the option to "Include curated regulatory domains" in the Association rule settings area of the input screen.
Currently, the curated regulatory domains are:
Curated Regulatory Domains used in GREAT version 4
We use the same set of curated regulatory domains as in the previous versions. The specific coordinates of the curated regulatory domains in each genome assembly are listed in the following files.
Human (hg19)
Sonic hedgehog long-range enhancer
- SHH: chr7:155438203-156584569
HOXD global control region
- LNP: chr2:176714855-176947690
- EVX2: chr2:176714855-176953690
- HOXD13: chr2:176714855-176959529
- HOXD12: chr2:176714855-176967083
- HOXD11: chr2:176714855-176976491
- HOXD10: chr2:176714855-176982491
Beta-globin locus control region
- HBB: chr11:5226931-5314124
- HBD: chr11:5253302-5314124
- HBG1: chr11:5260859-5314124
- HBE1: chr11:5276088-5314124
Human (hg18)
Sonic hedgehog long-range enhancer
- SHH: chr7:155130964-156277330
HOXD global control region
- LNP: chr2:176423101-176655936
- EVX2: chr2:176423101-176661936
- HOXD13: chr2:176423101-176667775
- HOXD12: chr2:176423101-176675329
- HOXD11: chr2:176423101-176684737
- HOXD10: chr2:176423101-176690737
Beta-globin locus control region
- HBB: chr11:5183507-5270700
- HBG1: chr11:5209878-5270700
- HBE1: chr11:5232664-5270700
Mouse (mm9)
Sonic hedgehog long-range enhancer
- Shh: chr5:28644270-29642207
Hoxd global control region
- Lnp chr2:73770001-74496641
- Evx2 chr2:74284768-74502220
- Hoxd13 chr2:74284768-74508266
- Hoxd12 chr2:74284768-74515449
- Hoxd11 chr2:74284768-74525027
- Hoxd10 chr2:74284768-74531027
Beta-globin locus control region
- Hbb-b1 chr7:110945345-111023716
- Hbb-b1 chr7:110967438-111023716
- Hbb-bh1 chr7:110981442-111023716
- Hbb-y chr7:110996677-111023716